Today we are officially announcing the relaunch of Do the Woo as a fully dedicated WooCommerce podcast under the Open Channels network. The show returns with a new mission, a refined focus, and a commitment to becoming the central place where the entire WooCommerce ecosystem can learn, share, and connect. More information about the relaunch …
Category: Dinoflagellate genomics
Dec 16
Open Channels FM: Do the Woo Podcast is Back with Conversations for Builders, Merchants, and Developers
The “Do the Woo” podcast is returning, focusing on WooCommerce insights, featuring industry leaders. It will include lively discussions, audience Q&As, and monthly episodes with hosts James Kemp and Katie Keith.
Dec 16
Introducing WPFilters: The Easiest Way to Add Advanced Search Filtering to WordPress
Ever looked at the search filters on Amazon or Airbnb and wished you could make the content on your own website just as easily discoverable? Imagine if your visitors could simply click a few options in the search filter, like “Price: Low to High,” “Rating: 4 Stars,” or “Color: Blue”… and instantly, the perfect results …
Dec 16
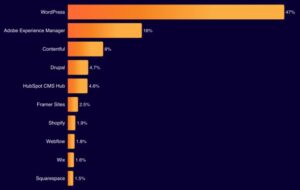
Matt: Cloudflare CMS Stats
Cloudflare released their Radar report for 2025, one fun stat was they analyzed the top 5,000 domains, and it had some interesting results for website technologies. Open Source tech came in at 47% for WordPress and 4.7% for Drupal, which wins the majority! Then in proprietary there was Adobe at 16%, Contentful at 8%, Shopify …
Dec 15
7 Best WooCommerce Sliding Side Cart Plugins (I Tested 13)
Cart abandonment is one of the most frustrating challenges for any online store owner. You’ve done the hard work to attract shoppers, but many still leave before completing their purchases. One common reason? Shoppers get annoyed when they have to leave the product page just to check what’s in their cart. That’s where sliding side …
Dec 15
Matt: Nex Playground
The Wall Street Journal has a fun article about the Nex Playground, The Hottest Toy of the Year Is Made by a Tech Startup You’ve Never Heard Of. I think this is the first time one of Audrey Capital’s companies (we invested when it was the Homecourt app) is the hot Christmas item. Here’s it …
Dec 14
Matt: Apple iWeb
The new X/Twitter algorithim is hard to predict, but I’ve had one go viral with over a million views now, a quote-tweet of a cool demo video of Apple’s website builder from 2009, with themes and blog support and everything. Interesting to compare its interface to Gutenberg and WordPress today. We don’t even know how …
Dec 13
Matt: Winter Fun
The colors here have now gone blue for winter, and snow has started, thanks to the excellent Snow Fall plugin. I also wanted to congratulate Wealthfront on their IPO. Many on their team have been friends or advisors over the years, from David Fortunato responding to my email about their WordPress blog being on an …
Dec 12
I Tried 7 Best GoFundMe Alternatives (Raise More, Pay Less)
You shouldn’t have to lose control of your donor data and rely on a third-party platform just to create a fundraising campaign. But that’s exactly what happens when you hit ‘launch’ on GoFundMe. Thankfully, there are plenty of GoFundMe alternatives out there. Some let you maintain 100% control over your donor relationships, others offer a …
Dec 12
Matt: Aldeas
Tonight was a lot of threads connecting for me. At Automattic’s Noho Space we hosted an event for Martin Scorsese’s new documentary about Pope Francis, called Aldeas. There was a point in my life when I wanted to become a priest, and I had been inspired by meeting a Franciscan seminary student. I took it …